Engineering of Chlamydomonas reinhardtii to produce betalain pigments and the use of riboswitches to direct metabolic flux
This project aims to engineer Chlamydomonas reinhardtii to produce betalain pigments, using riboswitches to direct metabolic flux.
The Idea
Betalain pigments are plant metabolites showing a wide variety of potential applications in the pharmaceutical, agricultural and food industry. Betalains are currently used as food and cosmetic colorants [1], and can act as potent antioxidant [2], anti-cancer [3, 4], anti-lipidemic [5, 6] and anti-microbial molecules [7]. Certain intermediates of the betalain metabolic pathway also have pharmacological properties e.g. L-DOPA is used for the treatment of Parkinson’s disease [8]. Given these benefits, it is important to further understand the biosynthetic processes leading to betalain production and accumulation in plants and how this knowledge can be used to engineer new industrial production hosts using model organisms like microalgae.
The betalain pathway consists of three main enzymatic steps starting with the molecule tyrosine (Fig 1) [9]. The first step of the pathway displays an interesting enzymatic redundancy, where at least three cytochrome P450 proteins (CYP76AD1, 5 and 6) are able to perform the conversion of tyrosine to L-DOPA [10, 11]. CYP76AD1 can also catalyse the second reaction of the pathway, from L-DOPA to cyclo-DOPA, necessary for production of red coloration [12]. One molecule of cyclo-DOPA can then spontaneously condense with a molecule of betalamic acid, product of the DODA gene in the third step of the pathway [13], to give the visible red pigments, betacyanins. Alternatively, betalamic acid can also condense with amino groups to form the fluorescent yellow pigments, betaxanthins.
Figure 1. Betalain metabolic pathway
Different CYP76AD enzymes are important for controlling the types of betalains produced and thus the resulting colour balance from pink to yellow via intermediate hues. Transient transformation of Nicotiana benthamiana with CYP76AD1 and the rest of the betalain biosynthetic machinery results predominantly in betacyanin production (red colour), whereas when CYP76AD6 is substituted for CYP76AD1, plants only produce betaxanthins (yellow colour) [10]. Orange colours in betalain-pigmented species have been shown to result from the presence of both betacyanins and betaxanthins [14]. Silencing of CYP76AD1 expression in beet has been shown to decrease betacyanin content and increase betaxanthins [12], indicating that modulating CYP76AD1 expression should influence the proportions of these two types of betalain pigments. Altogether, these findings suggest CYP76AD enzymes could have an important regulatory role in determining pigment composition and final coloration. The simplicity of the betalain pathway, the redundancy observed in the first step of betalain biosynthesis and the fact that betalamic acid acts as a shared chromophore for the two final pigment types, makes the betalain pathway an ideal system for visual characterisation of enzymes and their significance for metabolic flux towards either branches of this pathway.
We therefore propose to heterologously express the betalain biosynthetic pathway in Chlamydomonas reinhardtii with metabolic flux artificially regulated towards betacyanin or betaxanthin production. This regulation is possible by the use of characterised riboswitches that respond to exogenous supplementation of culture media with thiamine and hydroxyethyl thiazole (HET). Such riboswitch-driven regulation of the branch point enzymes will allow the ratio of cyclo-DOPA to betalamic acid to be “controlled” and thereby selectively shift the overall coloration produced towards the red or the yellow spectra respectively. The ultimate aim of this project is to engineer a strain of Chlamydomonas reinhardtii that not only will be able to produce betalain pigments but also will be susceptible to external fine tuning towards a desired color mix resulting in particular hues.
The Team
Mr Alfonso Timoneda,
Graduate Student, Department of Plant Sciences, University of Cambridge
Dr Payam Mehrshahi,
Postdoctoral scientist, Department of Plant Sciences, University of Cambridge
Dr Samuel Brockington,
Research group leader, Department of Plant Sciences, University of Cambridge
Project Outputs
Project Report
Summary of the project's achievements and future plans
Project Proposal
Original proposal and application
Project Resources
Progress Report: 1st March 2018
Summary
Betalain pigments are plant metabolites showing a wide variety of potential applications in the pharmaceutical, agricultural and food industry. Given the potential shown, it is important to further understand the biosynthetic processes leading to betalain production and accumulation in plants and how this knowledge can be used to engineer new industrial production hosts using model organisms like microalgae. We proposed to heterologously express the betalain biosynthetic pathway in Chlamydomonas reinhardtii with metabolic flux artificially regulated towards betacyanin or betaxanthin production by the use of riboswitches that respond to exogenous supplementation of culture media with thiamine and hydroxyethyl thiazole (HET). Such riboswitch-driven regulation of the branch point enzymes would allow the ratio of cyclo-DOPA to betalamic acid to be “controlled” and thereby selectively shift the overall coloration produced towards the red or the yellow spectra respectively. We have synthesised the sequences of five betalain biosynthetic genes (CYP76AD1, CYP76AD6, DODAa, and cDOPA5GT) previously domesticated and optimised to reflect codon usage in the nuclear genome of C. reinhardtii. We also isolated two recently published constitutive promoter and terminator combinations that constitute new tools for gene expression in C. reinhardtii (RPL23, FDX1). Five different multigene vectors were successfully assembled via MoClo Golden Gate cloning and transformed in the nuclear genome of C. reinhardtii. Selection and confirmation of expressing clones will be done in the near future and further analysis will determine riboswitch ability to modulate the betalain pathway.
Report and outcomes
To produce C. reinhardtii strains that are able to produce betalains in a regulated manner, the following milestones had to be addressed.
Codon optimisation and gene synthesis
We synthesised the sequences of five betalain biosynthetic genes (CYP76AD1, CYP76AD6, DODAa, and cDOPA5GT) previously optimised to reflect codon usage in the nuclear genome of C. reinhardtii. Beta vulgaris transcript sequences were used as templates for the codon optimisation for all genes, except for cDOPA5GT which was retrieved from the Mirabilis jalapa transcriptome. Domestication was done against BpiI, BsaI and SapI restriction sites to ensure compatibility with MoClo and Loop Golden Gate assembly protocols. Synthesis, domestication and codon optimisation were performed by the GeneArt gene synthesis service provided by Thermo Fisher Scientific. In all cases, genes were synthesised with flanking regions containing compatible MoClo Golden Gate overhangs and BsaI restriction sites, and entry plasmids were generated including a kanamycin resistance gene. Plasmid design was performed in order to be able to be used directly as MoClo level 0 vectors.
See Table 1 for complete list of generated vectors.
Cloning of RPL23 and FDX1 promoter and terminator pairs
Shortly after starting the OpenPlant project, a report was published identifying new genic flanking sequences for improved transgenic expression in C. reinhardtii [1]. Initially, our list of available constitutive promoters for Chlamydomonas consisted of the well characterised HSP70:RBCS2 and PSAD. The amount of genes we were considering for our final multigene constructs and the low number of promoter/terminator pairs available translated into a considerable risk of silencing associated with the use of repetitive elements. We therefore aimed to isolate the recently characterised promoter and terminator sequences for the RPL23, RPL35 and FDX1 genes from C. reinhardtii genomic DNA. Primer design included the overhangs and BpiI restriction sites required for cloning into MoClo level 0 acceptors. Promoters were amplified using two different primer pairs, generating amplicons with either TACT or AATG 3’ overhangs, for their use as promoter only or promoter plus 5’UTR parts respectively (Figure 1). Amplification was successful for promoter/terminator pairs of RPL23 and FDX1, and was followed by gel extraction and cloning to corresponding level 0 acceptors. This was enough to complete our multigene construct design and further attempts to isolate RPL35 flanking sites were not performed, but can be done in the future.
See Table 1 for complete list of generated vectors.
Multigene vector construction
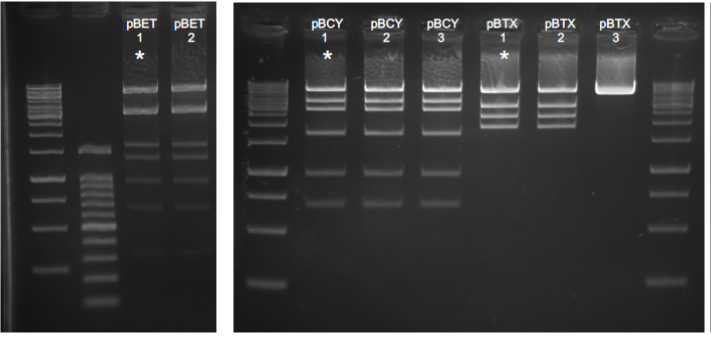
Using the MoClo Golden Gate technology we assembled multigene cassettes for their use in transformation of the Chlamydomonas nuclear genome. Final vector design is detailed in Table 2 at the end of the document, and new level 1 cassettes were constructed accordingly. Due to the relatively high number of genes included in some of the level 2 vectors, these were designed to contain two antibiotic resistance genes (for bleomycin and paromomycin) flanking both sides of the constructs. Dual selection would therefore ensure introduction of both ends of the cassette in the genome and minimize the number of selected colonies with loss of cassette fragments. The use of riboswitch parts in some of the level 1 cassettes decreased the assembly efficiency possibly due to its three-dimensional structures. We therefore used commercial NEB5a stable competent cells. Level 2 assemblies also proved themselves challenging probably due to vector complexity and had to be attempted through continuous repetitions and the optimization of part concentrations in some cases. All level 1 plasmids were checked by sequencing, and level 2 plasmids were checked by digestion with restriction enzymes (Figure 2) and sequencing of the cassette flanking regions.
See Table 1 for complete list of generated vectors.
Transformation of Chlamydomonas reinhardtii
C. reinhardtii strain uvm4 [2] has already been transformed by electroporation with the developed vectors. Transformants will undergo selection by growth in TAP media containing zeocin and paromomycin.
Strain characterization and riboswitch activation and tuning
After transformation of C. reinhardtii the following scenarios can be encountered:
(5.1) Pigmented colonies appear.
Gene presence and expression levels will be assessed by PCR and qPCR. Betalain production and composition in transgenic C. reinhardtii lines will be analysed using a spectrophotometer by measuring absorbance of betacyanins and betaxanthins at 540nm and 475nm respectively. Experiments to assess riboswitch ability to direct metabolic flux to either branch of the betalain pathway will be performed once the transgenic lines have been identified.
(5.2) Transformed C. reinhardtii strains don’t produce any coloration.
Gene presence on resistant colonies can be assessed by PCR to confirm integration of all the parts of our multigene cassette. In the cases where all genes are present, expression levels can be quantified by qPCR to rule out silencing of one or several of the transgenes. If these are as expected, new constructs with tagged versions of our transgenes should be designed to confirm protein presence and localisation.
(5.3) No colonies survive the selection process.
Transformation of C. reinhardtii would be repeated to rule out possible experimental errors. It is possible that one or more intermediates of the betalain pathway, or even pigment concentration itself, could have toxic effects on microalgae. To account for this possibility, transformation of C. reinhardtii with our riboswitch containing vector could be repeated, but this time adding thiamine to the selection media. This would inactivate the pathway hence allowing for a normal development of resistant colonies. Later, the colonies would be subcultured into liquid media containing a reduced concentration of thiamine that allows basal level of gene expression and thereby allow the optimization of expression ratios that would overcome potential cytotoxicity.
Figure 1. PCR performed to isolate 5’ and 3’ flanking regions for RPL23, RPL35 and FDX1 from C. reinhardtii genomic DNA. A, promoter region using primers with 3’ overhang TACT; B, promoter region using primers with 3’ overhang AATG; Ter, terminator region. White squares indicate right sized bands.
Figure 2. Diagnostic digestions for level 2 vectors. Numbers correspond to different plasmid extracted from independent colonies. pBET and pBCY were digested with NheI. pBTX was digested with EcoRI. Asterisks correspond to correct expected digestion patterns.
Table 1. Complete list of generated level 0, level 1 and level 2 vectors compatible with the MoClo Golden Gate cloning technology.
Table 2. Detailed level 2 design for each of the constructed vectors. pBET is the vector containing riboswitches for CYP76AD1 and CYP76AD6. pBCY and pBTX are vectors designed to produce betacyanins (red) and betaxanthins (yellow) respectively.
Follow on plans
Following the selection process, betalain producing C. reinhardtii strains will be characterized and riboswitch efficacy to control protein ratios and ultimately the direction of metabolic flux towards red or yellow coloration will be assessed.
In order to facilitate the downstream analysis of the betalain compounds produced, extra funds provided would contribute to the purchase of a ZORBAX Eclipse XDB-C18 (5mm, 4.6 x 50 mm) column for its use in liquid chromatography (LC-MS) quantification and analysis.
The isolated FDX1 terminator contained an undomesticated BpiI site which interfered with the correct assembly of Level 2 cassettes. Extra funding could also be used for the synthesis of this part and RPL35 promoter and terminator regions in the case further cloning attempts were unsuccessful.
ZORBEX Eclipse XDB-C18 (5mm , 4.6 x 50 mm) – currently £257.55 in the University MarketPlace.
References
[1] López-Paz C, Liu D, Geng S and Umen JG. 2017. Identification of Chlamydomonas reinhardtii endogenous genic flanking sequences for improved transgene expression. Plant J, 92: 1232–1244.
[2] Neupert J, Karcher D, Bock R. 2009. Generation of Chlamydomonas strains that efficiently express nuclear transgenes. The Plant Journal, 57: 1140–1150.