Project Proposal: The use of synthetic biology tools to define the roles of LysM receptor-like kinases in legumes and cereals
The Idea
Plant roots are exposed to myriads of microbes including both potential pathogens and symbionts. Fungal cell wall components, including chitin oligosaccharides (COs), activate PAMP-triggered immunity (PTI), following binding to LysM receptor-like kinases. Symbiotic microorganisms also produce the chitin-based signals lipochitooligosaccharides (LCOs) that are also recognized by LysM receptors to induce symbiotic processes. This raises questions regarding how plants use structurally similar receptors to distinguish similar microbial signals. Our proposal aims to understand this process using genetic complementation and domain swaps between CO8 and Nod factor receptors in legumes and non-legumes, facilitated by synthetic biology tools.
Who We Are
JIC: Feng Feng , feng.feng@jic.ac.uk Feng Feng received his Ph.D. in plant biology from Tsinghua University, China in 2012. After completion of his degree, he worked as a research assistant for one year in plant immunity at the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences. He then shifted his academic focus towards symbiosis signalling and began his postdoctoral work in Prof. Giles Oldroyd’s Laboratory at the John Innes Centre in December, where he has been since 2013. He is part of the Bill and Melinda Gates Foundation-funded programme that has pioneered the use of synthetic biology strategies in biotechnology.
Cambridge: Ronelle Roth, rr472@cam.ac.uk Ronelle Roth is a research associate in the group of Dr Uta Paszkowski and works on arbuscular mycorrhiza symbiosis (AMS) in the model crop species rice. Her project involves investigating the role of mycorrhiza-induced receptor kinases during AMS and pioneering 3-D live cell imaging and quantitative modelling of membrane dynamics during arbuscule development. She obtained her DPhil from Oxford University under the guidance of Prof Jane Langdale working on cellular differentiation during maize leaf development. She subsequently did post-doctoral research working on plant-pathogen interactions in the group of Prof Pierre de Wit at Wageningen University, The Netherlands. She was awarded a Marie Curie FP7-PEOPLE-IEF Fellowship in 2014 to work on beneficial plant-fungal interactions and this enabled her to return to science following a career break.
Implementation
In the natural environment plants are exposed to huge numbers of microbes, including potential symbionts and pathogens. Among beneficial interactions between microbes and plants are the associations between legumes and nitrogen-fixing rhizobium bacteria and the interactions between most species of plants and arbuscular mycorrhizal fungi (AMF). These interactions are promoted following plant recognition of diffusible signals from these microbes. The nodulation factors (Nod factors) and mycorrhizal factors (Myc factors) are chitin-based lipochitooligosaccharides (LCOs) or simple chitooligosaccharides (COs). These are perceived by plant LysM receptor-like kinases to initiate the common symbiosis signalling pathway, that promotes root nodule formation and mycorrhization (Oldroyd, 2013)
Plants also recognise signals from potential pathogens via pattern recognition receptors (PRRs) that sense pathogen-associated molecular patterns (PAMPs) to activate PAMP-triggered immunity (Jones & Dangl, 2006). Chitin is a major structural component of fungal cell walls, consisting of chitooligosaccharide backbone with β1-4 linked N-acetyl-D-glucosamine residues. The long chitooligosaccharides (COs) from fungi function as PAMPs and are recognized by a receptor complex to activate PTI signalling, that includes the production of reactive oxygen species (ROS), MAPK activation and pathogen growth inhibition (Jones & Dangl, 2006).
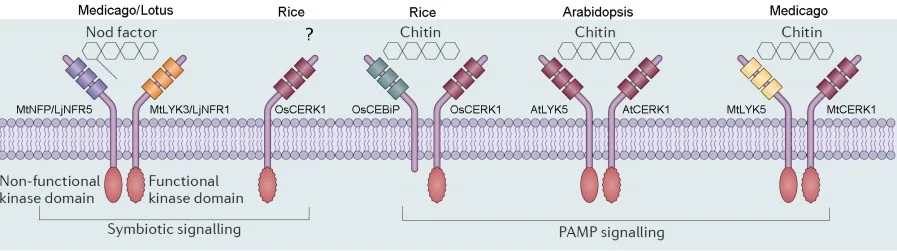
Plant Lysin-motif containing receptor-like kinases (LysM RLKs) mediate recognition of chitin-related molecules to activate defence and symbiosis signalling (Fig 1). In Arabidopsis thaliana, two LysM receptor genes AtCERK1 and AtLYK5 are required for CO8-induced PTI. The LysM domains of both receptors directly bind CO8, AtLYK5 having a much higher binding affinity. After chitin elicitation, AtCERK1 interacts with AtLYK5 to activate downstream signalling. Since AtLYK5 has no kinase activity, the kinase domain of AtCERK1 is likely responsible for chitin signal transduction (Liu et al., 2012; Cao et al., 2014). In rice (Oryza sativa), the chitin-binding protein (CEBiP) has extracellular LysM domains but no intracellular kinase domain. The LysM domain of this protein can also bind chitin directly and associate with a LysM RLK OsCERK1 to initiate defence signalling (Shimizu et al., 2010). Genetic and biochemical studies in the model legumes Lotus japonicus and Medicago truncatula have identified two LysM RLK genes LjNFR1/MtLYK3 and LjNFR5/MtNFP that are required for rhizobia Nod factor perception and symbiosis signalling. It has been shown that the kinase inactive receptor LjNFR5/MtNFP associates with LjNFR1/MtLYK3 that have a functional kinase domain, constituting the Nod factor receptor complex (Broghammer et al., 2012; Moling et al., 2014). The receptors in legumes associated with the perception of Myc factors are still unknown, however, it was found that OsCREK1 is required for mycorrhization in rice, and its close homolog MtLYK3 and LjNFR1 also plays a role in mycorrhizal infection (Zhang et al., 2015).
Figure 1: The LysM receptors are responsible for the recognition of chitin and Nod factor in plants
From the above recognition model (Fig 1), we know that the LysM domains of different receptors are responsible for the binding and recognition of different chitin-based ligands. It is possible that the exchange of LysM domains between different receptors could transfer specificity of recognition. This has already been shown for some receptors: using domain swaps it has been demonstrated that expression of chimeric receptors are a useful way to characterise ligand recognition and signal transduction. Wang et al designed chimeric constructs replacing the ectodomain of AtCERK1 in Arabidopsis with the ectodomain of LjNFR1 and LjNFR5 from Lotus. The resultant chimeric receptors transferred the ability to recognise Nod factors to Arabidopsis. Furthermore, the substitution of ectodomains of LjNFR1 and LjNFR5 with the corresponding region of OsCERK1 and OsCEBiP also switched the recognition specificity from Nod factor to chitin (Wang et al.,2014). Therefore, replacement of ectodomains of LysM RLKs provides an effective tool to understand the specificity and interaction between receptors and ligands. However, the construction of such chimeric receptors is laborious using traditional cloning techniques.
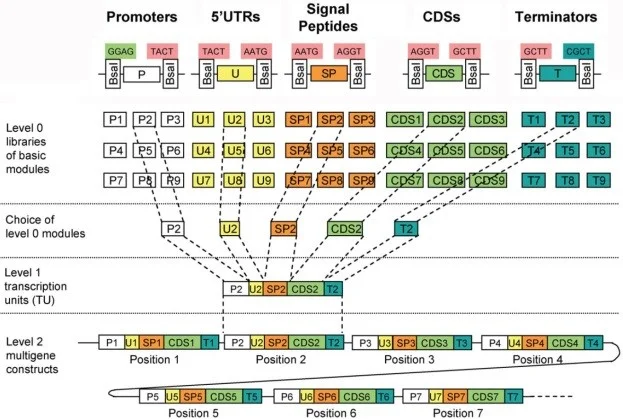
Golden gate cloning allows the rapid and flexible assembly of DNA constructs, using a combinatorial approach from an array of genetic elements (Fig 2). This technology is based on the special ability of type IIS restriction enzymes to cleave outside of their recognition site, the cleaved products contain compatible sequence overhangs allowing DNA fragments to be easily ligated. This cloning strategy develops three levels of modular libraries. The level 0 modules consist of promoters, 5’ untranslated regions, signal peptides, coding sequences and terminators with each module flanked by specific fusion sites to allow formation of complete transcriptional units. The resultant level 1 modules can then be assembled into the destination vector, creating the final construct for transformation (Weber et al., 2011).
Figure 2: General overview of Golden gate cloning system
Aims and Methodology
Using reverse genetic screening approaches we have identified two LysM RLKs that are required for CO8-induced PTI in Medicago and sequence alignments showed that they are close homologs of AtLYK5 and AtCERK1, suggesting that these two genes are CO8 receptors in Medicago. The identification of CO8 and Nod factor receptors in Medicago gives us a unique opportunity to assess the signalling pathway specificity for activation of defence and symbiosis within the same species. In collaboration with Uta Paszkowski’s group in Cambridge University, we will use synthetic biology approaches to assess the signalling specificity of different LysM RLKs from Medicago and rice. The aims of the project are:
1.Characterization of CO8 receptors in Medicago and rice
We will assess the interchangability of Medicago and rice CO8 receptors. For this we will express MtLYK5 and MtCERK1 under the control of theOsCERK1 promoter in the rice oscerk1 mutant, and also express OsCERK1 under the control of the MtLYK5 or MtCERK1 promoters in the mtlyk5and mtcerk1 mutants of Medicago. We will assess complementation in these lines using ROS production and the activation of MAPKs following CO8 treatment. Because rice OsCERK1 also plays essential roles in AM colonization, it will be very interesting to test which the CO8 receptors ofMedicago can complement the mycorrhizal deficient phenotype of oscerk1. In order to undertake these transcomplementation experiments, we will create golden gate modules of the receptors and promoters to generate the transformation vectors.
2. Determination of receptor recognition specificities for CO8 and Nod factor
We will design a number of chimeric constructs with domain swaps and motif substitutions between CO8 receptors and Nod factor receptors ofMedicago in an attempt to transfer binding specificities between the receptors. We will exchange the LysM domains between Nod factor receptors MtNFP and MtLYK3 and CO8 receptors MtLYK5 and MtCERK1, and then complement the chimeric genes in these receptor mutants to detect defence responses or nodulation. For this we will create level 0 modules of the receptors with amino acid substitutions, as well as creating versions of the domains of the receptors for interchange.
The groups in Cambridge and Norwich will work closely together to design the level 0 modules for promoters and different versions of coding sequence of these receptors, and then combine other modules that we already have to build the Level 2 constructs for plant complementation. Our group will host the training of Cambridge collaborators for Golden gate cloning. Working together we will generate the constructs necessary and test these in the different species, using the rice expertise in Cambridge and the Medicago expertise in Norwich. We will share the protocols and modules for all experiments.
This project will use synthetic biology approaches to test important questions in receptor recognition. These results will help us to understand the evolution of LysM RLKs required for the recognition of similar signalling molecules from different species to trigger different signal transduction pathways. This will provide a better framework for transferring Nod factor recognition to cereals, an important criteria in the Gates-funded programme. Moreover, the Pazskowski group will have the first exposure to golden gate cloning through this collaboration and we will share all existing golden gate modules, as well as our expertise in this process.
Benefits and outcomes
Our proposal uses golden gate cloning to answer an important question, and this is a good example of the application of synthetic biology in research. Our group has good experience in this technology and a commitment to share our expertise. The project builds a new collaboration in OpenPlant, through integrating the Pazskowski group. All components coming from this project and the existing modules will be shared through our well established web-based servers. Both groups in the team have strong reputations in the symbiosis field, the Norwich group focused on the legume symbiosis, whereas the Cambridge group is focused on cereals. The communication coming through this collaboration, will extend the utility of synthetic biology tools, providing further complementary between Norwich and Cambridge.
Budget
Modules synthesis
Group Element Number Size Price (9p/bp) Total
JIC Promoter 4 2kb £720 £3960
Coding sequence 12 2kb around £2160
Cambridge Promoter 2 2kb £360
Coding sequence 4 2kb around £720